

Welcome! Click here to login or here to register.
| |
|||||||||||||||||||
|
|||||||||||||||||||
NHEJ in Homo sapiens
nonhomologous end-joining
| Proteins: |
|
|
||||||||||||
NHEJ in KEGG: hsa03450
[picture]
[information]
NHEJ in REACTOME: 73889
|
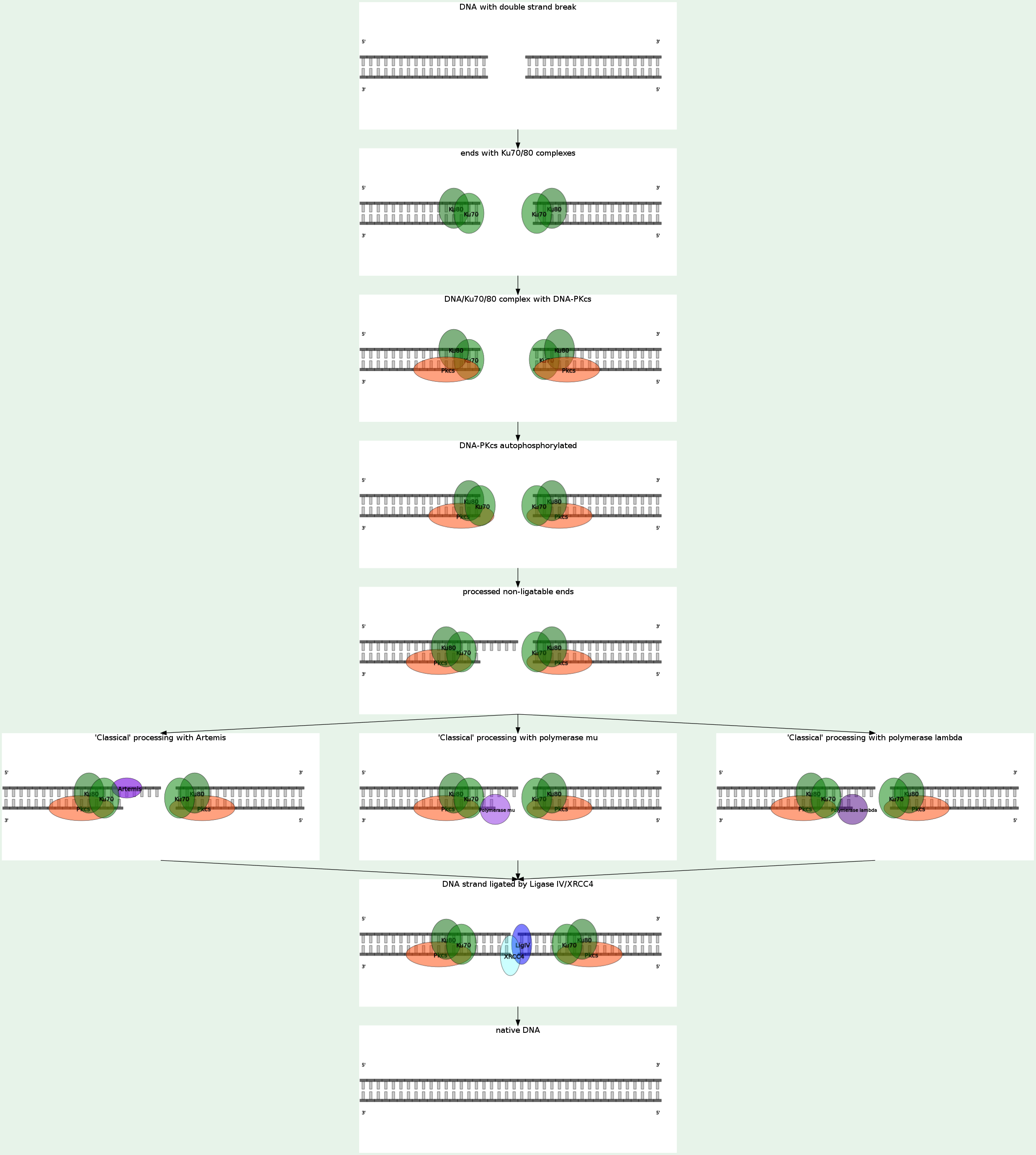
Nonhomologous end joining (NHEJ) eliminates DNA double-strand breaks (DSBs) by direct ligation. NHEJ involves binding of the KU heterodimer to double-stranded DNA ends, recruitment of DNA-PKcs (MRX complex in yeast), processing of ends, and recruitment of the DNA ligase IV (LIG4)-XRCC4 complex, which brings about ligation. A recent study shows that bacteria accomplish NHEJ using just two proteins (Ku and DNA ligase), whereas eukaryotes require many factors. NHEJ repairs DSBs at all stages of the cell cycle, bringing about the ligation of two DNA DSBs without the need for sequence homology, and so is error-prone.
NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homologous recombination, which requires a homologous sequence to guide repair. NHEJ typically utilizes short homologous DNA sequences called microhomologies to guide repair. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks of tumor cells. NHEJ is evolutionarily conserved throughout all kingdoms of life and is the predominant double-strand break repair pathway in mammalian cells. NHEJ in human is DNA-PK-dependent, KU70-KU80-dependent, LIG4-dependent. When the NHEJ pathway is inactivated, double-strand breaks can be repaired by a more error-prone pathway called microhomology-mediated end joining (MMEJ). In this pathway, end resection reveals short microhomologies on either side of the break, which are then aligned to guide repair. This contrasts with classical NHEJ, which typically uses microhomologies already exposed in single-stranded overhangs on the DSB ends. Repair by MMEJ therefore leads to deletion of the DNA sequence between the microhomologies. MMEJ in human is KU70-KU80-independent, DNA-PK-independent, LIG4-independent, LIG3-dependent. End binding and tethering The complex of MRE11-RAD50-NBS1 (MRN) is involved in NHEJ, but it may function at multiple steps in the pathway beyond simply holding the ends in proximity. DNA-PKcs is also thought to participate in end bridging during mammalian NHEJ. Eukaryotic Ku is a heterodimer consisting of Ku70 and Ku80, and forms a complex with DNA-PKcs, which is present in mammals but absent in yeast. Ku is a basket-shaped molecule that slides onto the DNA end and translocates inward. Ku may function as a docking site for other NHEJ proteins, and is known to interact with the DNA ligase IV complex and XLF. End processing End processing involves removal of damaged or mismatched nucleotides by nucleases and resynthesis by DNA polymerases. This step is not necessary if the ends are already compatible and have 3' hydroxyl and 5' phosphate termini. Little is known about the function of nucleases in NHEJ. Artemis is required for opening the hairpins that are formed on DNA ends during V(D)J recombination, a specific type of NHEJ, and may also participate in end trimming during general NHEJ. MRE11 has nuclease activity, but it seems to be involved in homologous recombination, not NHEJ. The X family DNA polymerases Pol λ and Pol μ fill gaps during NHEJ. Ligation The DNA ligase IV complex, consisting of the catalytic subunit DNA ligase IV and its cofactor XRCC4, performs the ligation step of repair. XLF, also known as Cernunnos, is also required for NHEJ. While the precise role of XLF is unknown, it interacts with the XRCC4/DNA ligase IV complex and likely participates in the ligation step. Recent evidence suggests that XLF re-adenylates DNA ligase IV after ligation, recharging the ligase and allowing it to catalyze a second ligation. |
Keywords:
Add your own comment!
There is no comment yet.
|
|

|

|