

Welcome! Click here to login or here to register.
| |
|||||||||||||||||||
|
|||||||||||||||||||
NHEJ in Saccharomyces cerevisiae
nonhomologous end-joining
| Proteins: |
|
|||||||||||
NHEJ in KEGG: sce03450
[picture]
[information]
|
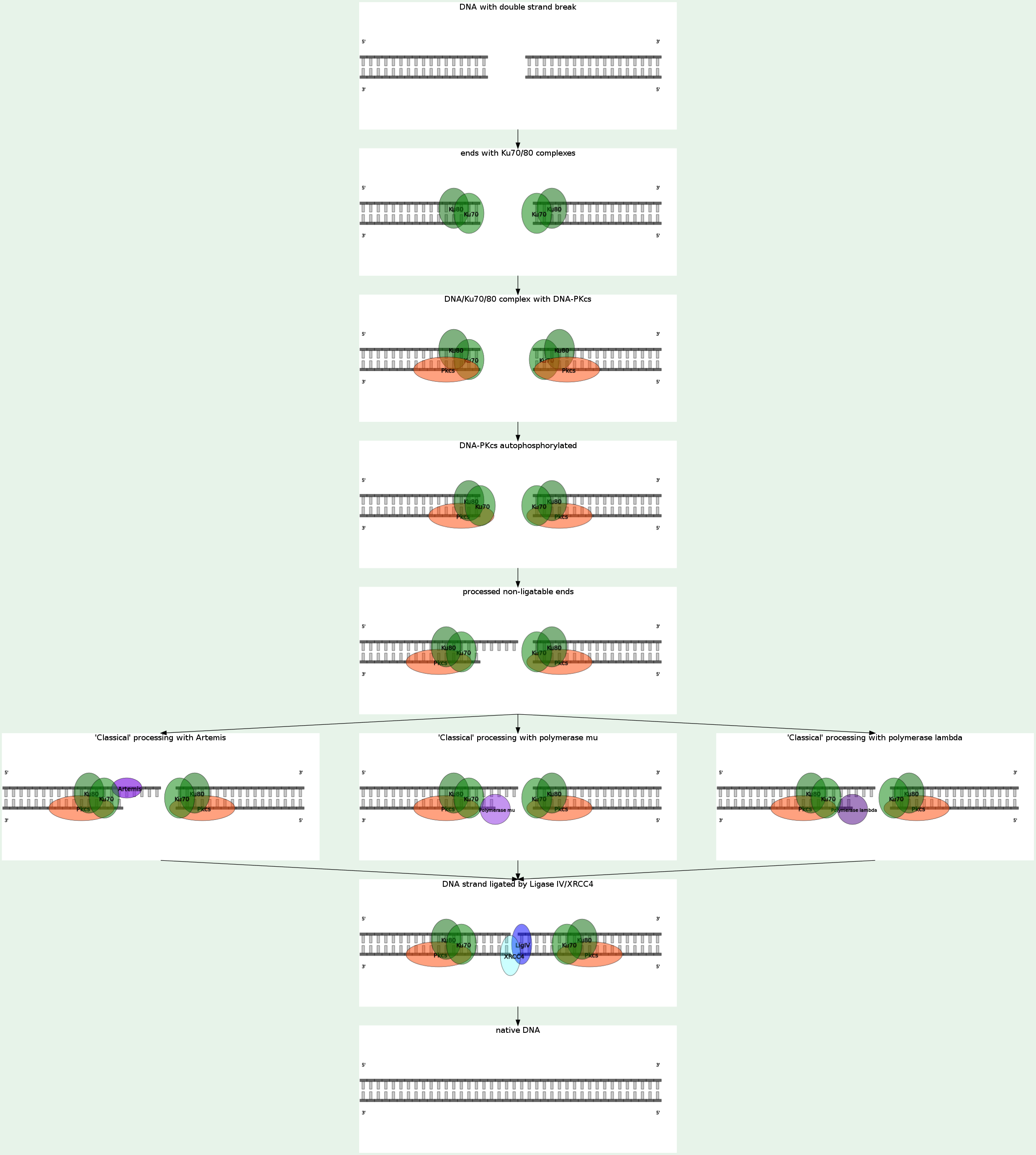
Nonhomologous end joining (NHEJ) eliminates DNA double-strand breaks (DSBs) by direct ligation. NHEJ involves binding of the KU heterodimer to double-stranded DNA ends, recruitment of DNA-PKcs (MRX complex in yeast), processing of ends, and recruitment of the DNA ligase IV (LIG4)-XRCC4 complex, which brings about ligation. A recent study shows that bacteria accomplish NHEJ using just two proteins (Ku and DNA ligase), whereas eukaryotes require many factors. NHEJ repairs DSBs at all stages of the cell cycle, bringing about the ligation of two DNA DSBs without the need for sequence homology, and so is error-prone.
NHEJ in yeast is Yku70p-dependent, Dnl4p-dependent, DNA-PKcs-independent (lack in yeast). When the NHEJ pathway is inactivated, double-strand breaks can be repaired by a more error-prone pathway called microhomology-mediated end joining (MMEJ). In this pathway, end resection reveals short microhomologies on either side of the break, which are then aligned to guide repair. This contrasts with classical NHEJ, which typically uses microhomologies already exposed in single-stranded overhangs on the DSB ends. Repair by MMEJ therefore leads to deletion of the DNA sequence between the microhomologies. MMEJ in yeast is Yku70p-independent, Dnl4p-independent, Nej1-dependent, Srs2-dependent, Sae2-dependent, Tel1-dependent. However, in budding yeast, homologous recombination (Rad52-dependent) dominates when the organism is grown under common laboratory conditions. End binding and tethering In yeast, the Mre11-Rad50-Xrs2 (MRX) complex is recruited to DSBs early and is thought to promote bridging of the DNA ends. Eukaryotic Ku is a heterodimer consisting of Ku70 and Ku80, and forms a complex with DNA-PKcs, which is present in mammals but absent in yeast. Ku is a basket-shaped molecule that slides onto the DNA end and translocates inward. Ku may function as a docking site for other NHEJ proteins, and is known to interact with the DNA ligase IV complex and XLF. End processing End processing involves removal of damaged or mismatched nucleotides by nucleases and resynthesis by DNA polymerases. This step is not necessary if the ends are already compatible and have 3' hydroxyl and 5' phosphate termini. Little is known about the function of nucleases in NHEJ. Mre11 has nuclease activity, but it seems to be involved in homologous recombination, not NHEJ. The X family DNA polymerase Pol4 in yeast fills gaps during NHEJ. Yeast lacking Pol4 are unable to join 3' overhangs that require gap filling, but remain proficient for gap filling at 5' overhangs. This is because the primer terminus used to initiate DNA synthesis is less stable at 3' overhangs, necessitating a specialized NHEJ polymerase. Ligation The DNA ligase IV complex, consisting of the catalytic subunit DNA ligase IV (Dnl4 in yeast) and its cofactor Lif1 (in yeast), performs the ligation step of repair. Nej1 is also required for NHEJ. DNA ligase IV after ligation, recharging the ligase and allowing it to catalyze a second ligation. Other In yeast, Sir2 was originally identified as an NHEJ protein, but is now known to be required for NHEJ only because it is required for the transcription of Nej1. |
Keywords:
There is no comment yet.
|
|

|

|