|
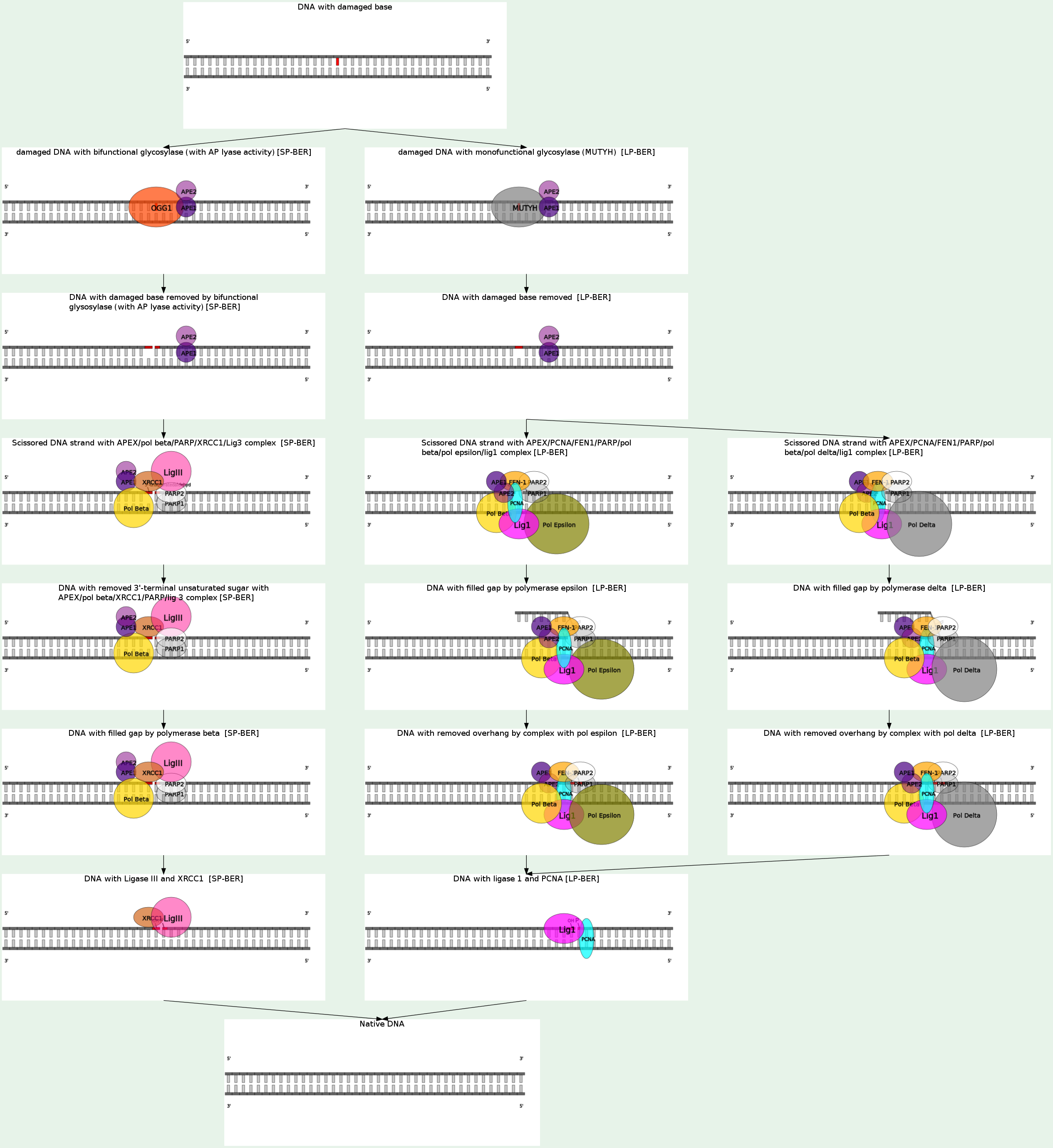
BER is the major repair pathway involved in the removal of DNA damage involving structurally non-distorting and non-bulky lesions, e.g. oxidized or ring-saturated bases, alkylated and deaminated bases, as well as apurinic/apyrimidinic sites, and also some type of mismatches.
Glycosylases are generally divided into two types:
1) monofunctional (Tag, AlkA, Ung, Mug and MutY) - remove a deaminated, alkylated or mismatched base leaving an AP-site
2) bifunctional (Fpg, Nth, and Nei) - remove oxidized bases and additionally to the glycosylase activity have a 3'-AP-lyase activity which incises the phosphodiester bond at the 3'-side of the deoxyribose via beta-elimination leaving a single strand break (SSB) with 3'-phospho-alpha, beta-unsaturated aldehyde (3'-PUA) and 5'-phosphate (5'P) ends. Moreover, Fpg and Nei bifunctional glycosylases additionally carry out beta-elimination reaction with removal of the deoxyribose residue and generation of 3'-phosphate termini (3'P).
The AP-sites or DNA ends generated after lesion excision or excision and incision by mono- or bifunctional glycosylases are not suitable for the next repair steps. AP-endonuclease (APE) is the main enzyme responsible for processing of the BER-intermediates. E. coli APEs, Xth (exonuclease III) and Nfo (endonuclease IV) efficiently incise AP sites and remove both beta- and beta/gamma-elimination products.
BER proceeds further via two alternative subpathways: short-patch (SP), which involves replacement of one nucleotide, or long-patch (LP), which involves replacement of several nucleotides (at least 2, often 6–13 nucleotides). In E. coli cells DNA polymerase I (DNA pol I) fills the gap and DNA ligase I seals the nicked DNA strands.
|