|
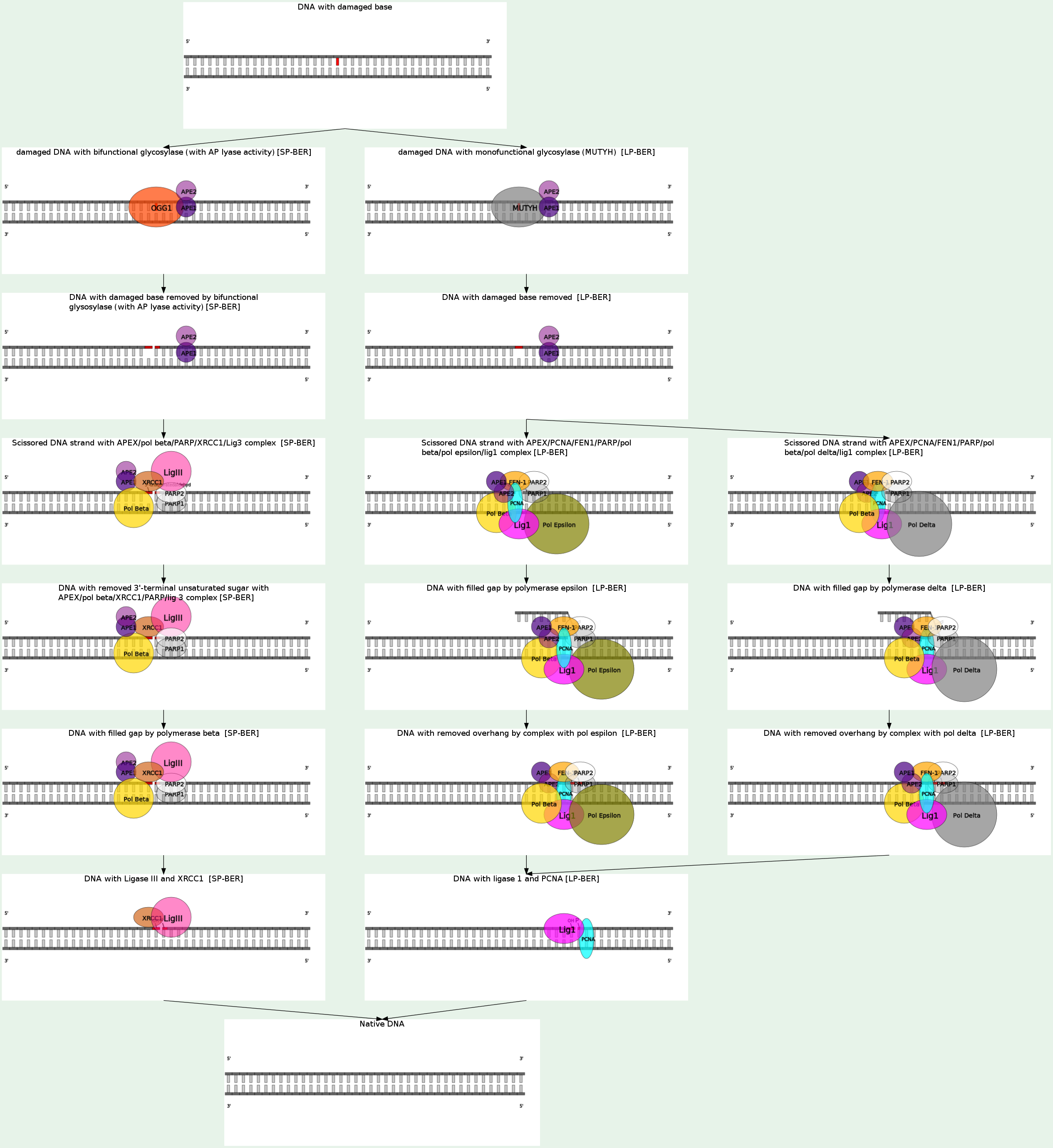
Base excision repair (BER) is a cellular mechanism that repairs damaged DNA throughout the cell cycle. It is primarily responsible for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch (where a single nucleotide is replaced) or long-patch BER (where 2-10 new nucleotides are synthesized). Single bases in DNA can be chemically damaged by a variety of mechanisms, most commonly deamination, oxidation, and alkylation. These modifications can affect the ability of the base to hydrogen-bond, resulting in incorrect base-pairing, and, as a consequence, mutations in the DNA. For example, incorporation of adenine across from 8-oxoguanine (right) during DNA replication causes a G:C base pair to be mutated to T:A. Other examples of base lesions repaired by BER include:
(i) oxidized bases: 8-oxoguanine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG, FapyA); (ii) alkylated bases: 3-methyladenine, 7-methylguanine; (iii) deaminated bases: hypoxanthine formed from deamination of adenine, tymine formed from deamination of 5-methylcytosine; (4) uracil inappropriately incorporated in DNA or formed by deamination of cytosine.
In addition to base lesions, the downstream steps of BER are also utilized to repair single-strand breaks. The choice between short- and long-patch repair is currently under investigation. Various factors are thought to influence this decision, including the type of lesion, the cell cycle stage, and whether the cell is terminally differentiated or actively dividing. Some lesions, such as oxidized or reduced AP sites, are resistant to pol β lyase activity and therefore must be processed by long-patch BER. Pathway preference may differ between organisms, as well. While human cells utilize both short- and long-patch BER, the yeast Saccharomyces cerevisiae was long thought to lack a short-patch pathway because it does not have homologs of several mammalian short-patch proteins, including pol β, DNA ligase III, XRCC1, and the kinase domain of PNKP. The recent discovery that the poly-A polymerase Trf4 possesses 5' dRP lyase activity has challenged this view.
|