|
NER is an important mechanism of DNA repair by which the cell can prevent unwanted mutations by removing the vast majority of UV-induced DNA damage (mostly in the form of thymine dimers and 6-4-photoproducts).
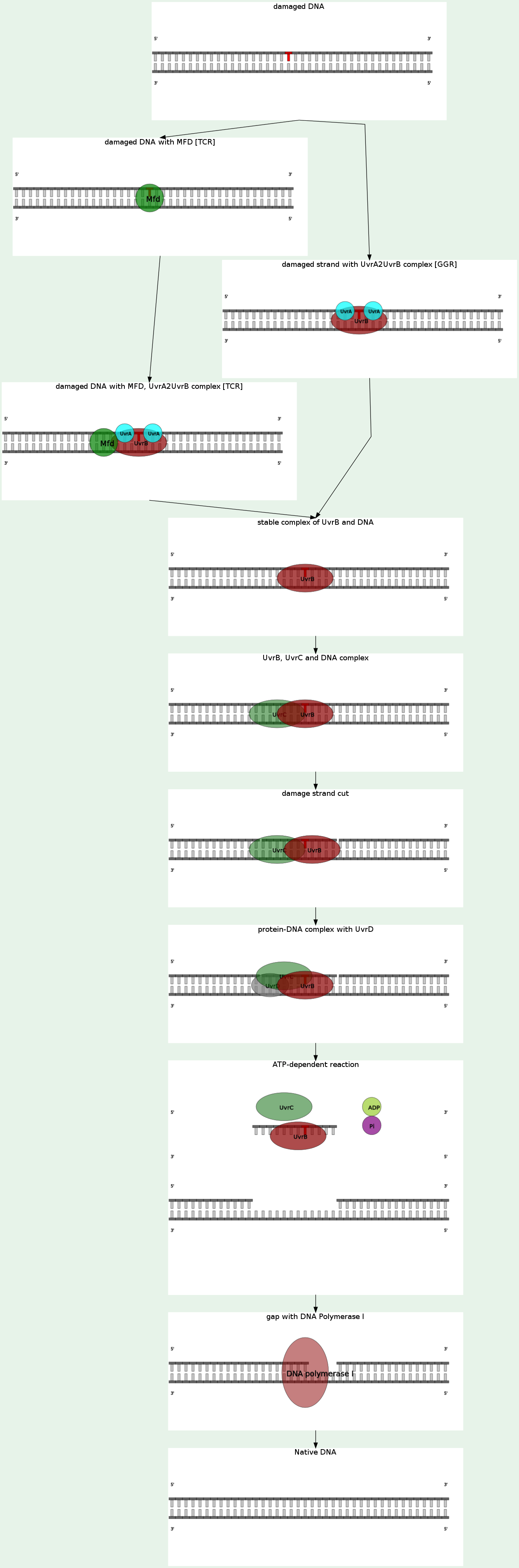
The process of nucleotide excision repair is controlled in E. coli by the UvrABC endonuclease enzyme complex, which consists of four Uvr proteins: UvrA, UvrB, UvrC, and DNA helicase II (sometimes also known as UvrD in this complex). First, a UvrA-UvrB complex scans the DNA, with the UvrA subunit recognizing distortions in the helix, caused for example by thymine dimers and other cyclobutyl dimers. When the complex recognizes such a distortion, UvrA exits and UvrB melts the base pairs between the two DNA strands. UvrB then recruits UvrC, which incises the DNA 8 nucleotides away from the distortion on the 5’ end and 4 nucleotides away on the 3’ end, creating a 12-nucleotide long single-stranded DNA that is excised with the help of the DNA helicase II. Next, the gap is filled in using DNA polymerase I and DNA ligase.
|